By Kieran D. Lamb et al.
Source Biorxiv
Abstract
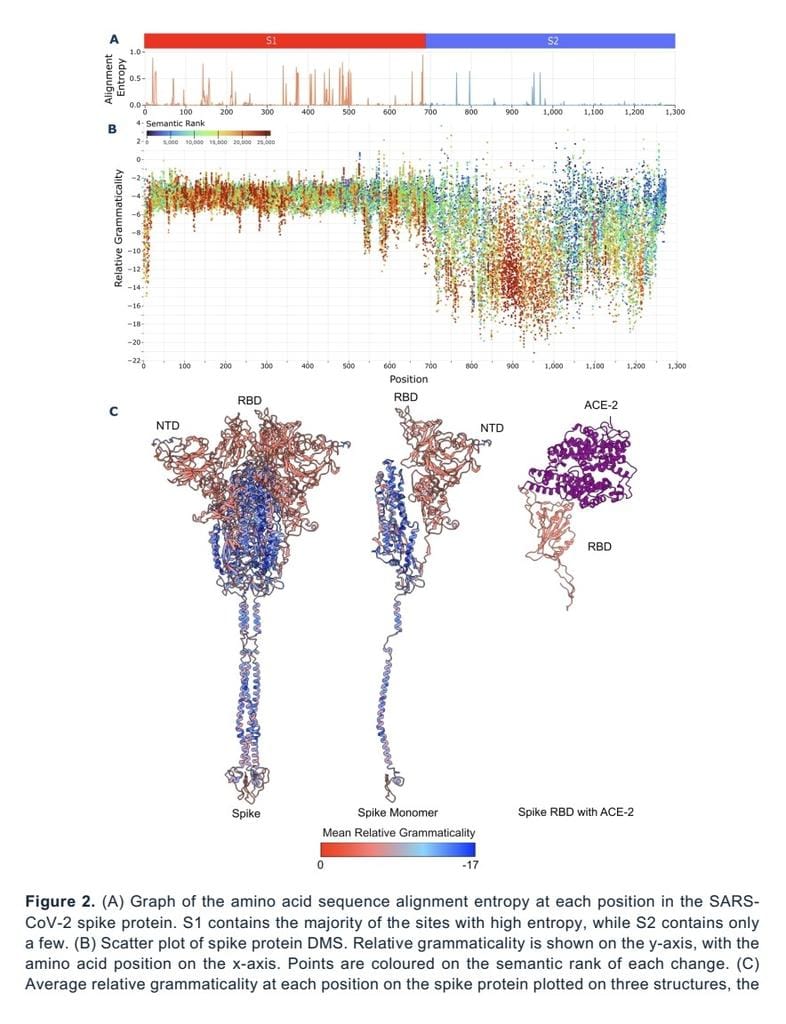
Protein language models (PLMs) capture features of protein three-dimensional structure from amino acid sequences alone, without requiring multiple sequence alignments (MSA). The concepts of grammar and semantics from natural language have the potential to capture functional properties of proteins. Here, we investigate how these representations enable assessment of variation due to mutation. Applied to SARS-CoV-2’s spike protein using in silico deep mutational scanning (DMS) we demonstrate the PLM, ESM-2, has learned the sequence context within which variation occurs, capturing evolutionary constraint. This recapitulates what conventionally requires MSA data to predict. Unlike other state-of-the-art methods which require protein structures or multiple sequences for training, we show what can be accomplished using an unmodified pretrained PLM. We demonstrate that the grammaticality and semantic scores represent novel metrics. Applied to SARS-CoV-2 variants across the pandemic we show that ESM-2 representations encode the evolutionary history between variants, as well as the distinct nature of variants of concern upon their emergence, associated with shifts in receptor binding and antigenicity. PLM likelihoods can also identify epistatic interactions among sites in the protein. Our results here affirm that PLMs are broadly useful for variant-effect prediction, including unobserved changes, and can be applied to understand novel viral pathogens with the potential to be applied to any protein sequence, pathogen or otherwise.
Read more click here